Crystallographic Structure of Druggable Enzymes from the Brain Parasite Toxoplasma
The twelve enzymes function in key metabolic pathways likely to be vital for parasite survival. Simply put, we starve the parasite!
Primary Collaborator: Wayne F. Anderson, PhD, Former Director of Center for Structural Genomics of Infectious Diseases, Chicago, IL.
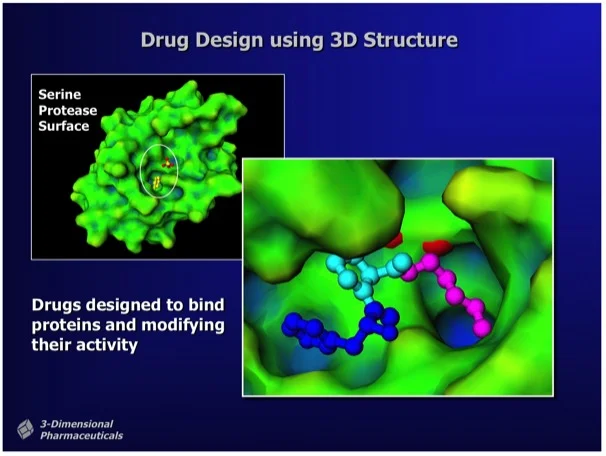
The mission is to develop a toolbox of drugs to combat the brain parasite Toxoplasma gondii. One approach is to use 3D protein structural data to aid in drug design (Figure 1).
These projects are open for all scientists to use (see BrainParasiter Journal, Entry 7, A passionate plea to my colleagues).
Please contact the current administration of CSGID for available reagents.
Note 1. Plasmodium is a 'kissing cousin' of Toxoplasma. They are evolutionary related and grouped together as the Apicomplexan parasites. This protozoan parasite causes malaria, a disease widespread in the tropical and subtropical regions. It causes ~750,000 deaths per year. A smart, but likely to be challenging, strategy is to develop and optimize drugs that disrupt the enzymes in both parasites with minimal effect on the human homologue.
Note 2. Tg in front of a gene name means Toxoplasma gondii. There is only one species gondii in the genus Toxoplasma, so I skip the formality of using both nomenclature and refer to the organism as Toxoplasma, or Toxo.
Note 3. Gene expression of parasite proteins during the life cycle of the brain parasite Toxo is interpreted from open-sourced data of ToxoDB.
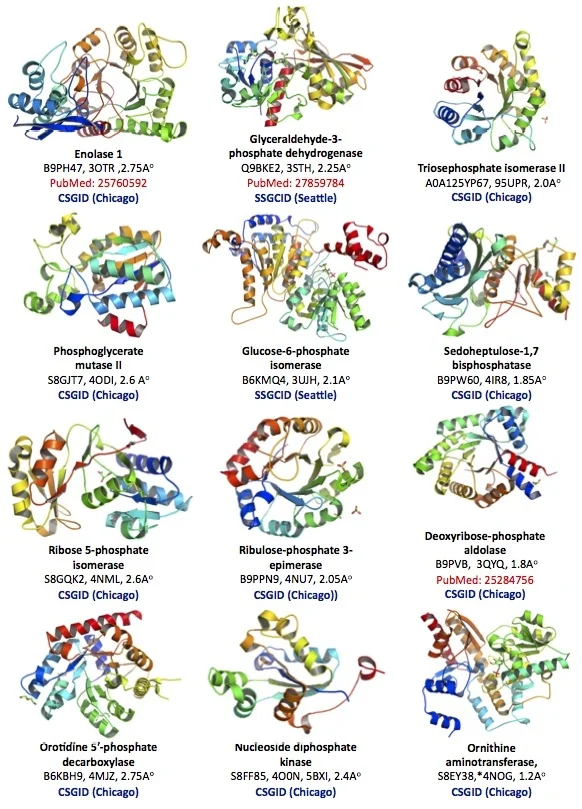
The protein structural database of the brain parasite Toxoplasma is poorly developed. Our goal is to contribute protein crystallographic structures of druggable small enzymes for the international research community to use (Figure 2).
Figure 1. "Structure-Based Drug Design uses the 3D structure of a drug lead bound to a target protein as a direct means to visualize drug-protein interactions and to determine chemical modifications that can improve potency." Ray Salemme's Website.
The twelve enzymes target key metabolic pathways likely to be vital for parasite survival. Simply put, we starve the parasite!
Most of these enzymes have homologues in humans. Most associate with more well-known human diseases, such as cancer and neuropathologies. The optimal strategy is to know well the parasite and human homologues so we can design structural-based therapy that hits two diseases with one ‘stone’, yet still prevents therapeutic side effects.
This structural-based strategy takes advantage of the large databases of protein structure (Protein Structure Data Bank) and over half a century of research in structural biology and drug design. It complements trendier cellular-based approaches, e.g. CRISPR gene editing, which need several decades to develop. Lessons taught from drug resistance of pathogens direct us to develop multiple drugs for the pipeline. We should consider the approach of a cocktail of different drugs, so many leads are needed.
These crystallographic structures also give opportunities to examine protein structure-functions in parasite biology, an underdeveloped field of study. Parasitologists are drawn to ‘novel’ features of Toxoplasma and consider metabolic enzymes as boring and unworthy. One missed opportunity is dissecting the fascinating multifunction of these ‘boring’ enzymes. In fact, most enzymes moonlight other important functions besides their metabolic role. Our published studies examined two such examples (GAPDH1, ENO1).
Figure 2. Crystallographic structure of 12 Toxoplasma enzymes deposited in the Protein Structural Data Bank (PDB).
Toxoplasma Enolase 1 (PDB#: 3OTR), Glyceraldehyde-3-phosphate dehydrogenase 1 (3STH), Triosephosphate isomerase II (95UPR), Phosphoglycerate mutase II (4ODI), Glucose-6-phosphate isomerase (3UJH), Sedoheptulose-1,7 bisphosphatase (4IR8), Ribose 5-phosphate isomerase, (4NML), Ribulose-phosphate 3-epimerase (4NU7), Deoxyribose-phosphate aldolase (3QYQ), Orotidine 5'-phosphate decarboxylase (4MJZ), Nucleoside diphosphate kinase (4O0N, 5BXI), Ornithine aminotransferase (5EAV, 5UPR, 4NOG, 4ZLV, 4ZWM, 5EQC, 5E3K, 5E5I, 5DJ9).
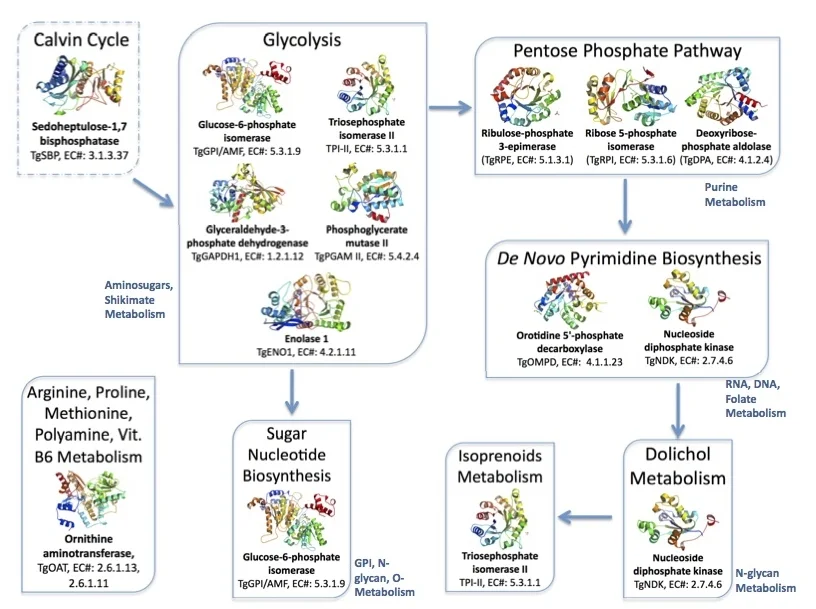
Figure 3. Metabolic pathways in the brain parasite Toxoplasma mediated by the twelve proteins with solved crystallographic structures.
Links: Calvin Cycle, Glycolysis, Pentose Phosphate Pathway, de novo Pyrimidine Biosynthesis, Dolichol, Isoprenoid Metabolism, Sugar Nucleotide Biosynthesis, and Ornithine aminotransferase.
The enzymes are summarized with our current knowledge and the directions needed for validation as drug targets. I plead to the research community to carry out these projects.
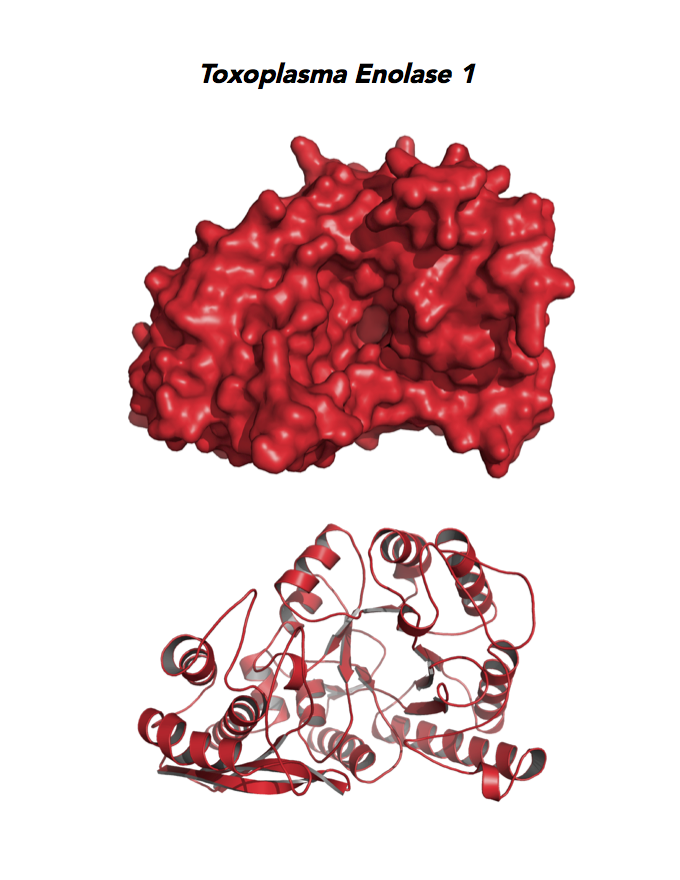
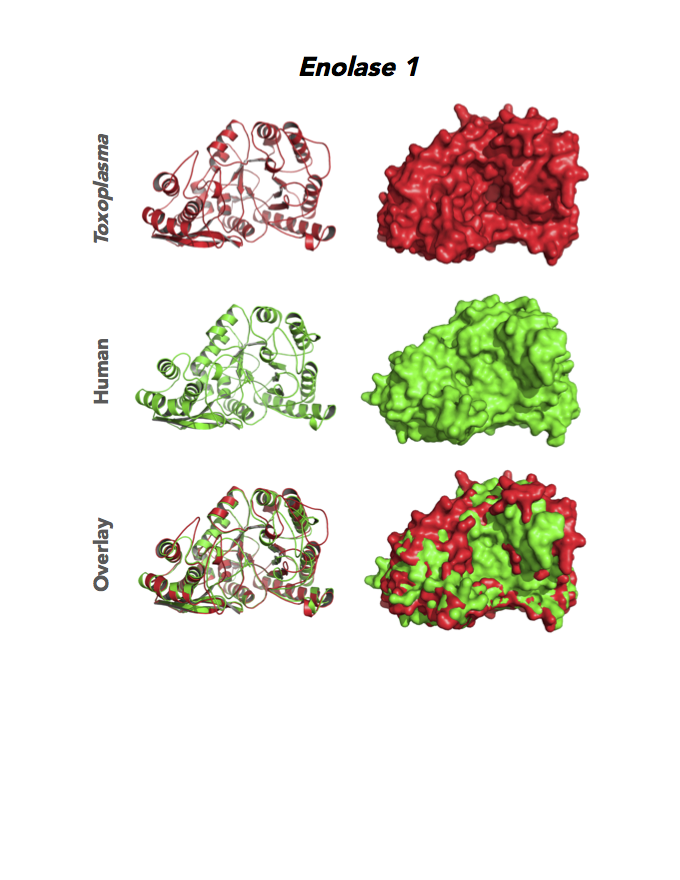
Target 1. Enolase 1 (TgENO1).
Enolase is a multifunctional protein. It catalyzes the ninth and penultimate step of glycolysis. An alternative-spliced isoform (MBP1) in humans binds to the c-myc promoter and functions as a tumor suppressor. Human ENO1 is a cell surface receptor for plasminogen during pathological conditions such as inflammation, autoimmunity, and malignancy.
Toxoplasma ENO1 expresses in the latent bradyzoite cyst stage, albeit with diminished catalytic activities due to a unique residue, Glu164, in catalytic loop 2. The isozyme has a surface positive charge that may promote binding to bradyzoite promoters. It regulates gene transcription in brain cyst development. A dual approach of drug targeting the parasite and cancer cells is best.
PMID: 25760592. Ruan et. al. 2005, Acta Crystallogr. D Biol. Crystallogr. 71:417.
PDB: 3OTR, 2.75Ao, UniProt: B9PH47, ToxoDB: TGME49_068860, EC#: 4.2.1.11, Molecular Function: Magnesium Ion Binding, Phosphopyruvate Hydratase Activity, Lyase Activity, Metal Ion Binding, Biological Process: Glycolytic Process.
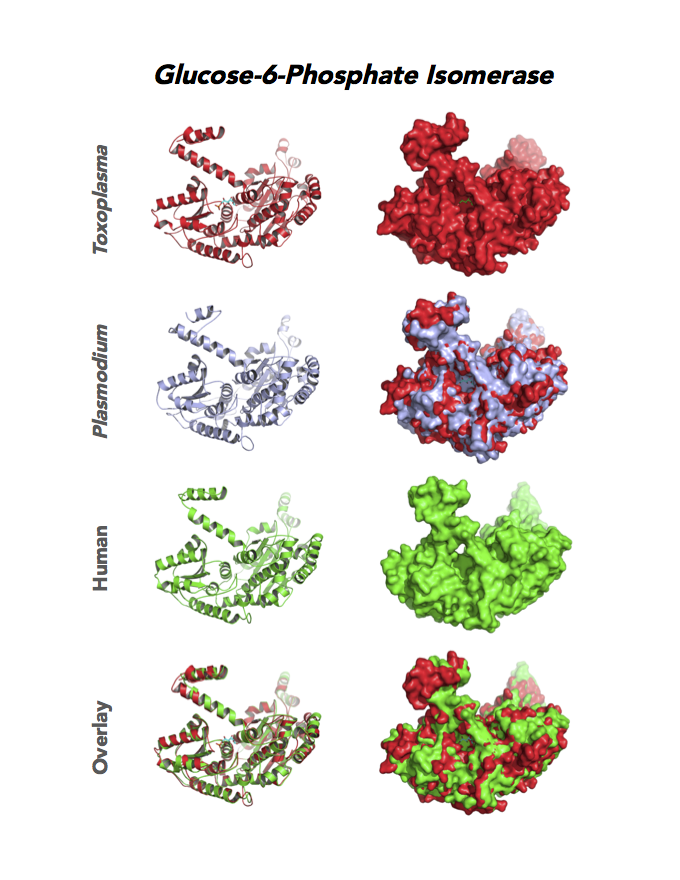
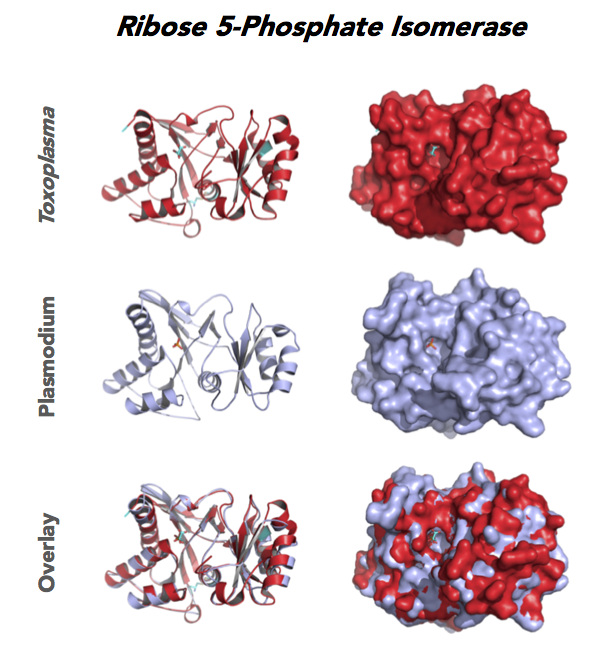
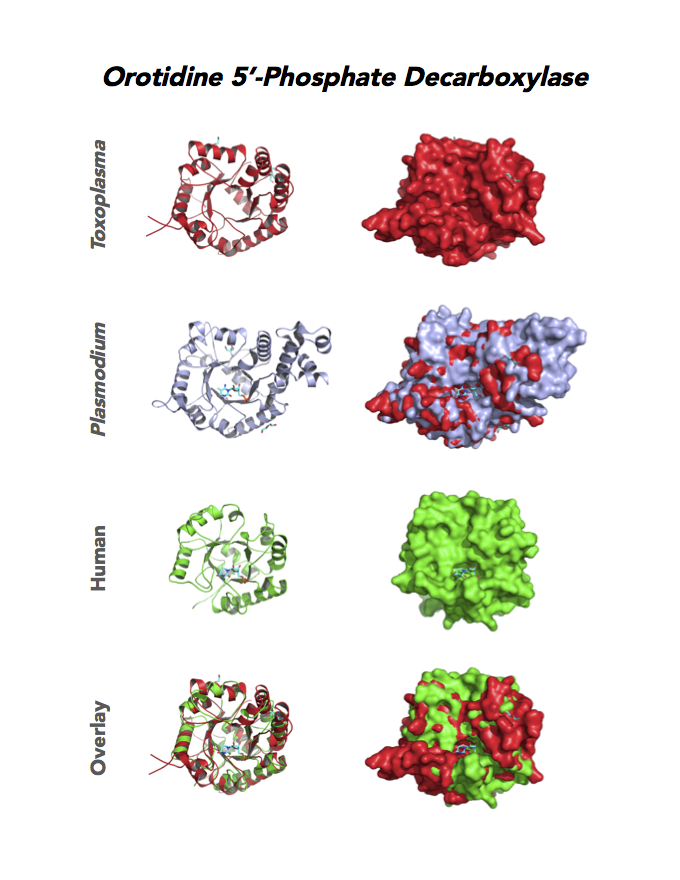
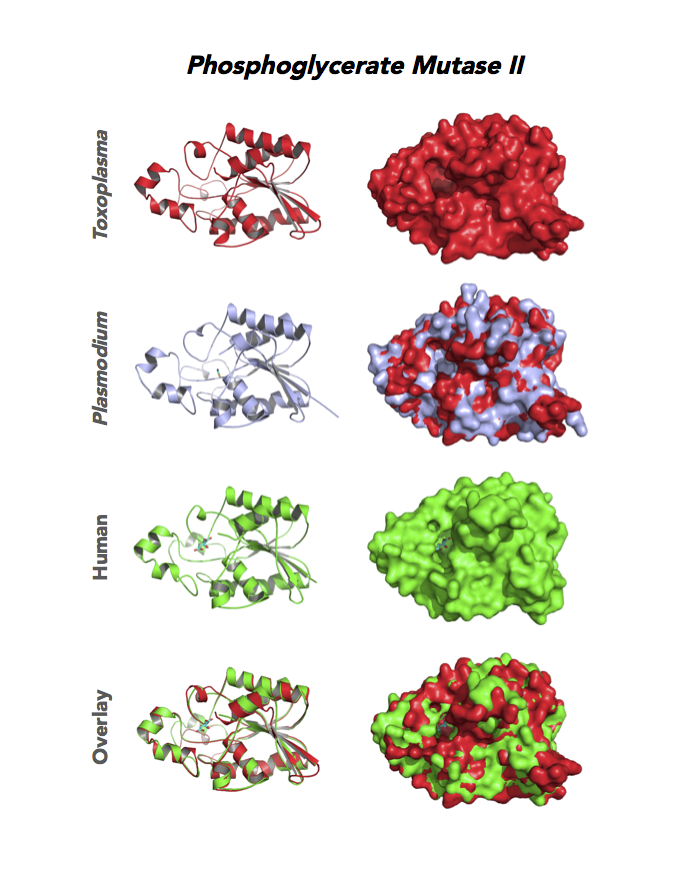
Figure 4. Four panels show crystallographic structures of Enolase 1 (ENO1) and Glucose-6-phosphate isomerase (GPI/AMF). First panel of each pair computes the rendering of enzyme surface (top) and skeleton of carbon backbone (bottom). Second panel compares the structure of enzymes from the brain parasite Toxoplasma (red), malaria Plasmodium (light blue), and human. Such comparison intructs the synthetic design of drugs that may inhibit parasite and not the human enzyme.
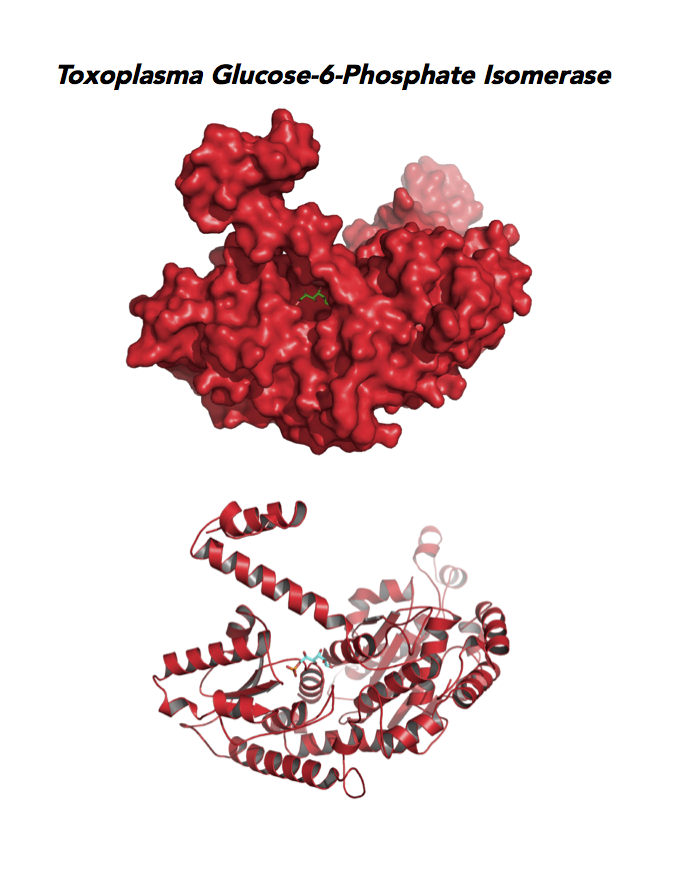
Target 2. Glucose-6-phosphate isomerase (TgGPI/AMF).
GPI is a multifunctional protein. Inside the cell, it takes part in glycolysis, gluconeogenesis and the pentose phosphate pathway. Outside the cell, it has significant mitogenic and cytokinic effects under its synonymous names as autocrine motility factor (AMF), neuroleukin agent (NLK), serine proteinase inhibitor, and differentiation and maturation mediator (DMM).
Due to its diverse and important role, GPI associates with severe enzyme deficiency disorder from genetic defects, autoimmune pathology in rheumatoid arthritis, and learning and memory in the central nervous system. PGI/AMF promotes the migration and invasion of cancer cells in tumor metastasis.
Toxoplasma RPI expresses in all life cycle stages, but this phosphohexose isomerase upregulates in the chronic phase of mice infection yielding bradyzoites and the post-sporulation of oocysts in cat intestinal wall. Protein expression derives from open-sourced data of ToxoDB. Its essentiality, metabolic and moonlighting functions are unknown. TgGPI structure allows comparison with human and Plasmodium GPI structures.
PDB: 3UJH, 2.1Ao, UniProt: A0A151H1D4, ToxoDB: TGME49_283780, EC#: 5.3.1.9, Molecular Function: Glucose 6 Phosphate Isomerase Activity, Biological Process: Gluconeogenesis, Glycolytic Process.
Plant Calvin Cycle.
We contributed crystallographic structures of five Toxoplasma enzymes likely evolved from the Calvin-Benson Cycle of plant plastid. They are TgGAPDH, TgRPE, TgRPI, TgTPI, and TgSBPase. Our collaborator solved a sixth enzyme structure (Fructose 1,6 bisphosphate aldolase, PDB# 4D2J, 4TU1) that is included in our joint publication. This pathway may be an alternate to make glucose in the parasite compared to human host. Several of these enzymes are not found in humans and might be exploited as parasite-specific chemotherapeutic targets.
Target 3. Glyceraldehyde-3-phosphate dehydrogenase 1 (TgGAPDH1)
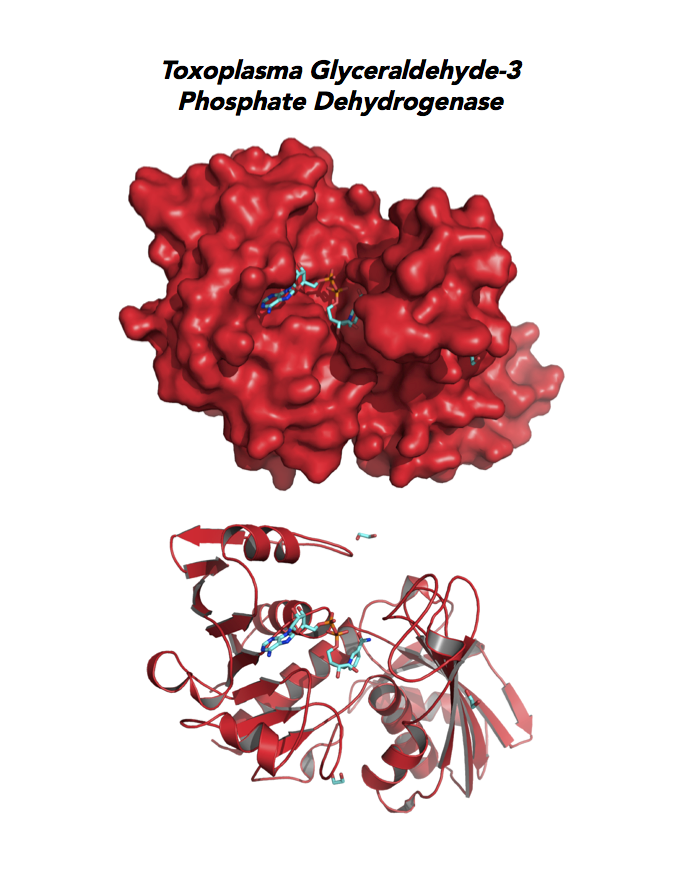
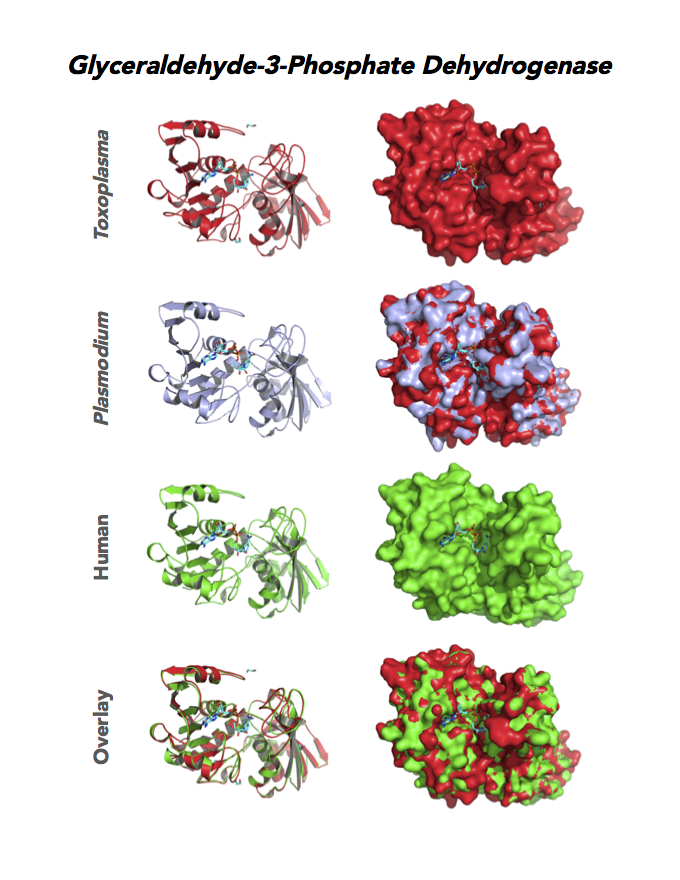
The enzyme GAPDH is the quintessential multifunctional protein. GAPDH catalyzes the sixth step of glycolysis. The protein functions in transcription activation, initiation of apoptosis, ER to Golgi vesicle shuttling, and fast axonal, or axoplasmic transport.
The dysfunction of GAPDH energy metabolism and other roles is implicated in multiple human cancers and neurodegenerative disorders.
In Toxoplasma, TgGAPDH1 is essential and expressed in every stage of its life cycle. Structure-function dissection reveals that phosphorylation of the S-loop mediates enzymatic activity. The attachment of GAPDH1 with the cortex is mediated by the N-terminus, likely palmitoylation. Drug design can target phosphorylation of residues S50 and S203 and Lys-Gly dipeptide insertion of the S-Loop. A joint approach of drug targeting the parasite, cancer cells and neurodegeneration is desirable.
PMID: 27859784. Dubey et al. 2017, Mol Microbiol. 103:618-634 (reprint).
PDB: 3STH, 2.25Ao, UniProt: Q9BKE2, ToxoDB: TGME49_089690, EC#: 1.2.1.12, Molecular Function: Glyceraldehyde 3 Phosphate Dehydrogenase Activity, Oxidoreductase Activity, NAD Binding, Biological Process: Glucose Metabolic Process, Glycolytic Process, Oxidation Reduction Process.
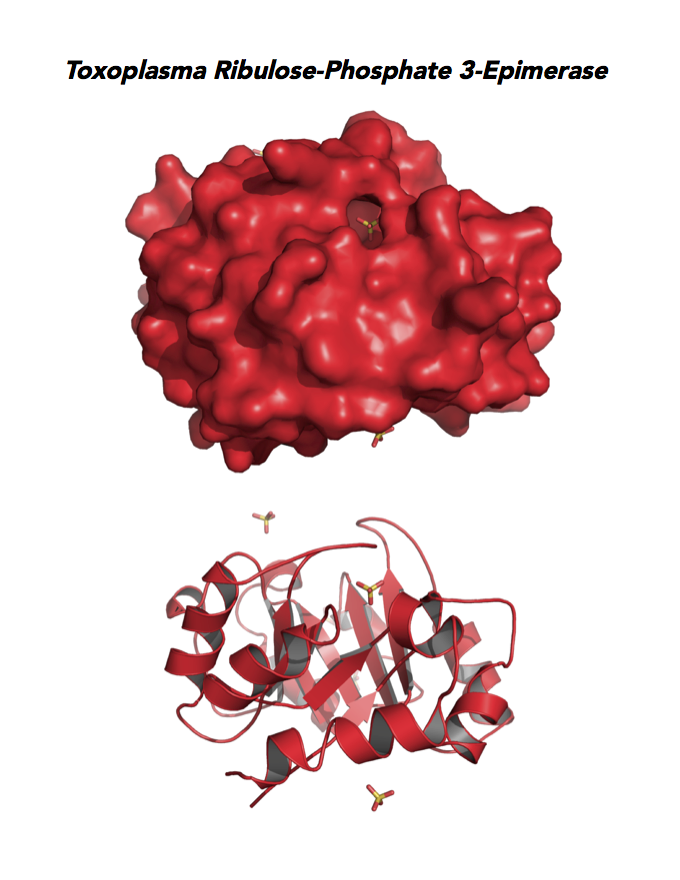
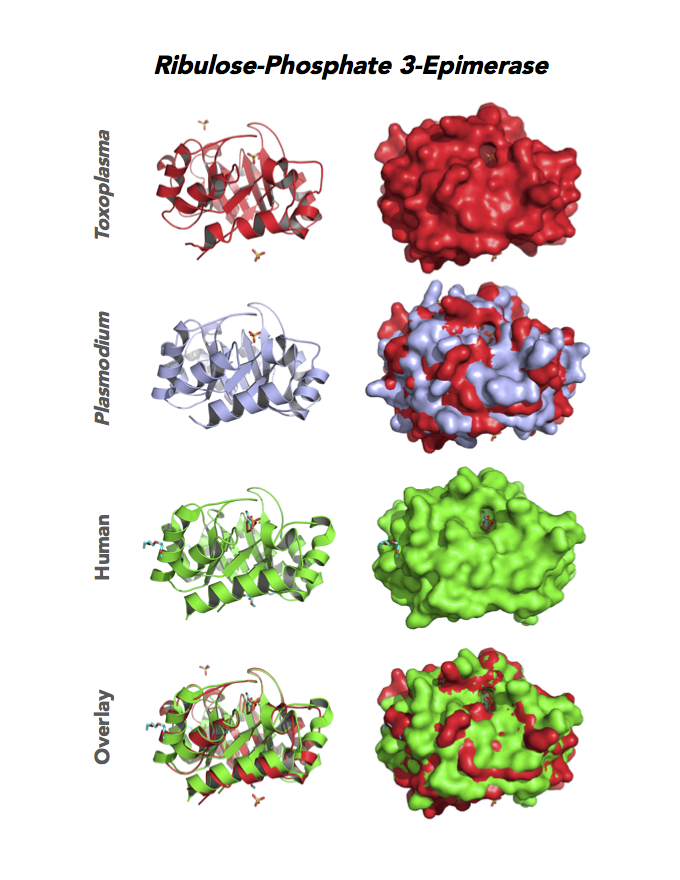
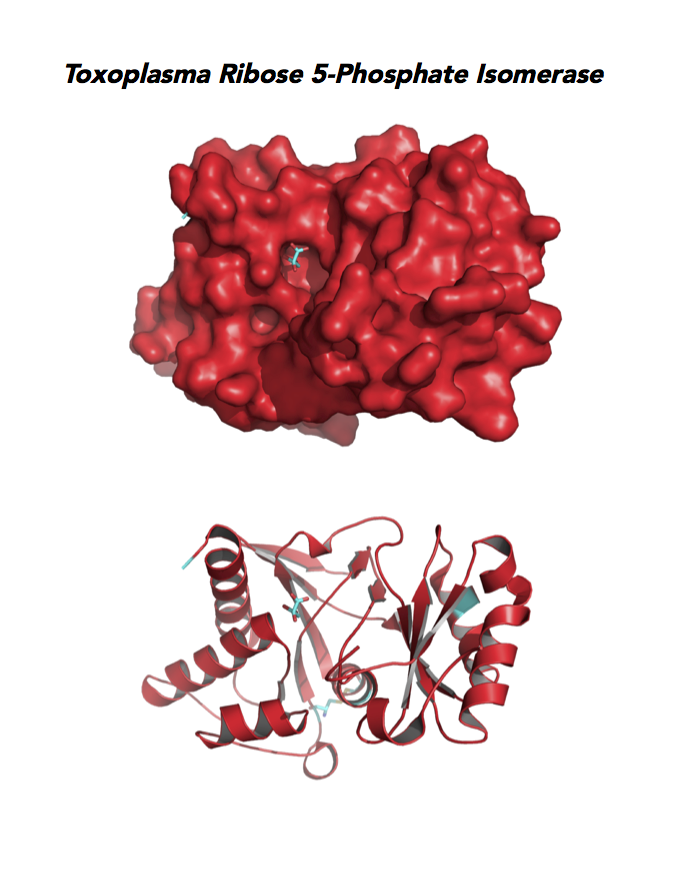
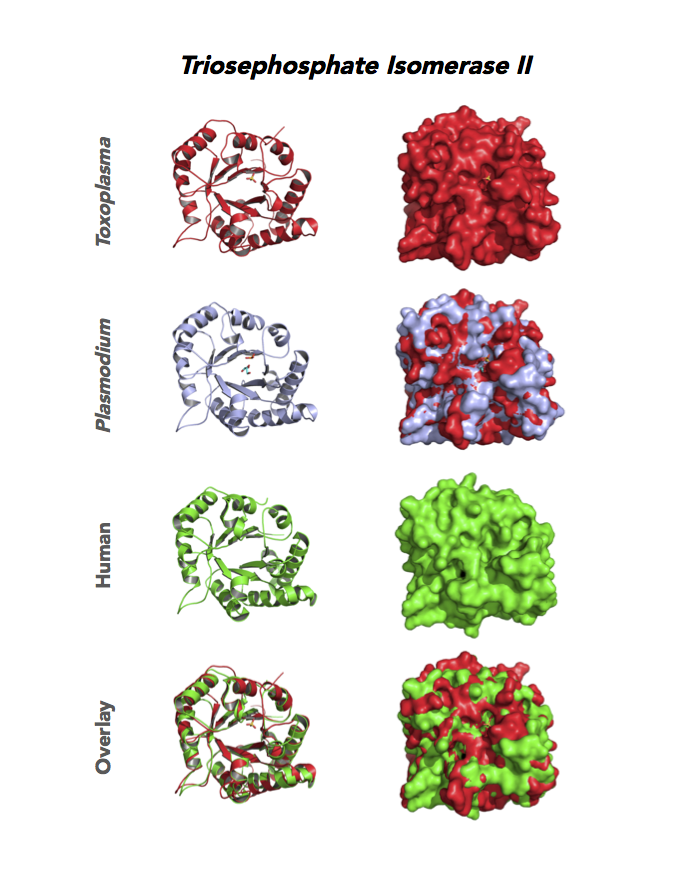
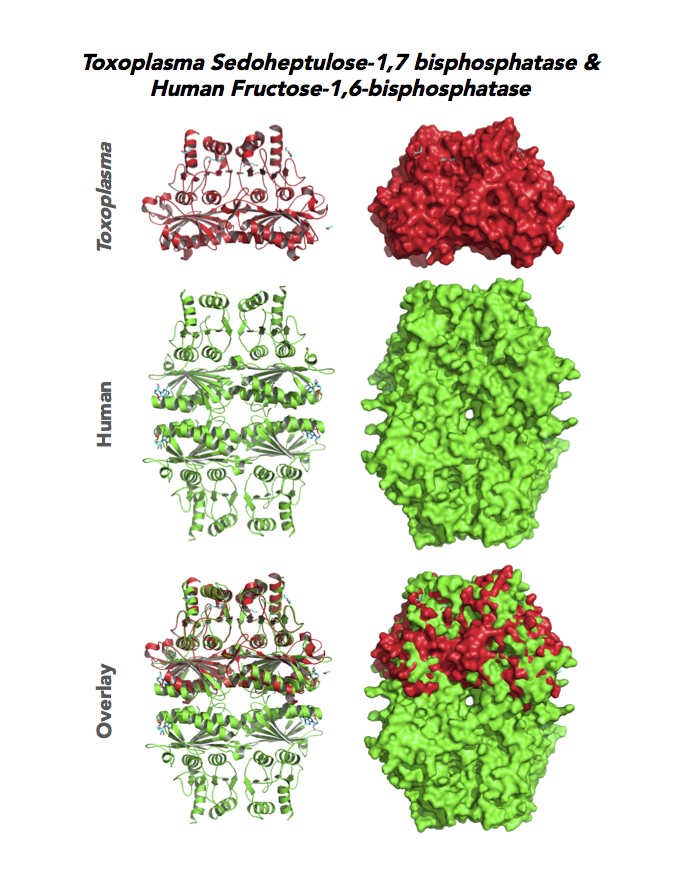
Figure 5. Ten panels show crystallographic structures of Calvin Cycle Enzymes. They are Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1), Ribulose 5-phosphate 3-epimerase (RPE), Ribose 5-Phosphate isomerase (RPI), Triosephosphate isomerase (TPI-II), and Sedoheptulose-1,7 bisphosphatase (SBP).
Target 4. Ribulose 5-phosphate 3-epimerase (TgRPE).
RPE is required for the Calvin cycle and for the non-oxidative phase of the pentose phosphate pathway. This metalloenzyme generates important precursors for production of energy, synthesis of aromatic amino acids, and protection against oxidative stress. RPE also yields precursors for conversion to chorismate from the shikimate pathway. Missing in animals, the shikimate pathway provides basic building blocks for the biosynthesis of folates and aromatic amino acids (phenylalanine, tyrosine, and tryptophan).
Toxoplasma RPE expresses in every stage of the parasite life cycle (ToxoDB), but the essentiality, metabolic and moonlighting functions are not known. TgRPE structure allows comparison with human and Plasmodium RPE structures.
PDB: 4NU7, 2.05Ao, UniProt: A0A0F7V7C1, ToxoDB: TGME49_047670, EC#: 5.1.3.1, Molecular Function: Catalytic Activity, Ribulose Phosphate 3 Epimerase Activity, Racemase and Epimerase Activity, Biological Process: Carbohydrate Metabolic Process, Pentose Phosphate Shunt, Metabolic Process.
Target 5. Ribose 5-Phosphate isomerase (TgRPI).
Type A of RPI is more common and the isozyme is found in most organisms, including human and Toxoplasma. Analysis in yeast and mammalian cells shows that RPI is essential for survival at the cellular level. It is the key regulator of the pentose phosphate pathway. RPI also plays a role in the control of autophagy.
RPIA affects several human diseases such as metabolic disorders and cancer. The rare genetic disease of RPI deficiency manifests with leucoencephalopathy and peripheral neuropathy. RPIA overexpression can induce oncogenesis in hepatocarcinogenesis. Modulation of RPIA levels in Drosophila suggests RPI might serve as therapeutic strategy for aging and neurodegenerative disorders.
Toxoplasma RPI is expressed in all life cycle stages (ToxoDB), but essentiality, metabolic and moonlighting functions have not examined. TgRPI structure allows comparison with Plasmodium RPE structures.
PDB: 4NML, 2.6Ao, UniProt: S8GQK2, ToxoDB: TGME49_239310, EC#: 5.3.1.6, Molecular Function: Ribose 5 Phosphate Isomerase Activity, Isomerase Activity, Biological Process: Pentose Phosphate Shunt Non Oxidative Branch.
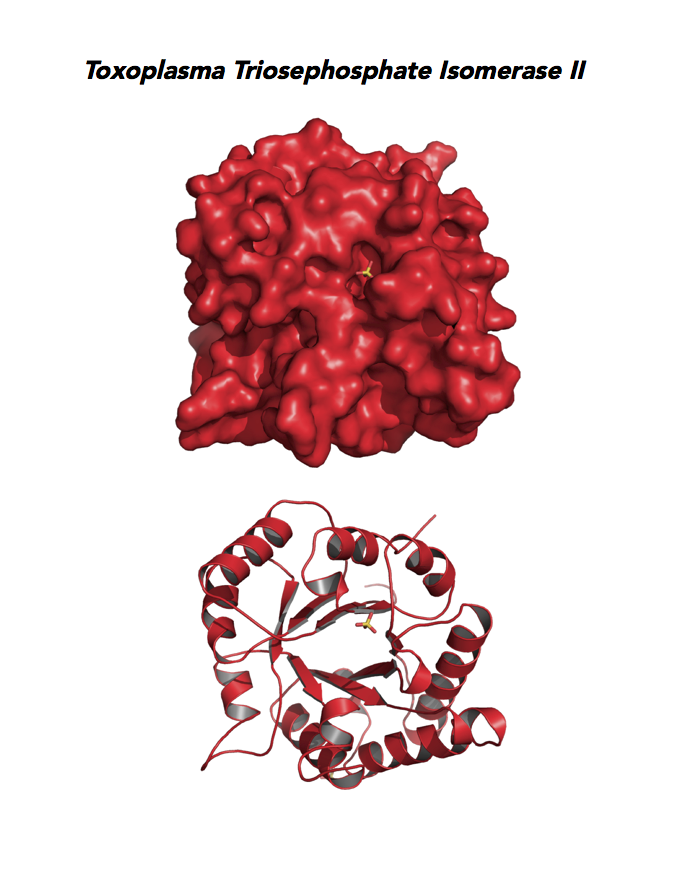
Target 6. Triosephosphate isomerase II (TPI-II).
TPI is in a metabolic crossroad that contributes to the glycerol synthesis pathway, glycolysis, the pentose-phosphate pathway, and the glycerol 3-phosphate NADH shuttle. This isomerase is one of the most studied enzymes yielding over 200 structures deposited in the PDB for 42 different species. Over 62% of protein residues have been examined by mutagenesis and/or using other approaches. TPI is a moonlighting protein in sperm–ZP glycoprotein interactions.
The enzyme is essential and plays a role in the pathology of Alzheimer’s disease, multiple cancers, and TPI Deficiency disorders.
Toxoplasma TPI-II localizes to the parasite residue plastid. Expression of TgTPI-II decreases in the chronic phase of mice infection and post-sporulation of oocysts (ToxoDB). Its essentiality, metabolic and moonlighting functions are not known. TgRPI structure allows comparison with the large repertoire of human and Plasmodium RPI structures.
PDB: 5UPR, 2.0Ao, UniProt: A0A125YP67, ToxoDB: TGME49_233500, EC#: 5.3.1.1, Molecular Function: Catalytic Activity, Triose Phosphate Isomerase Activity, Biological Process: Glycolytic Process, Metabolic Process.
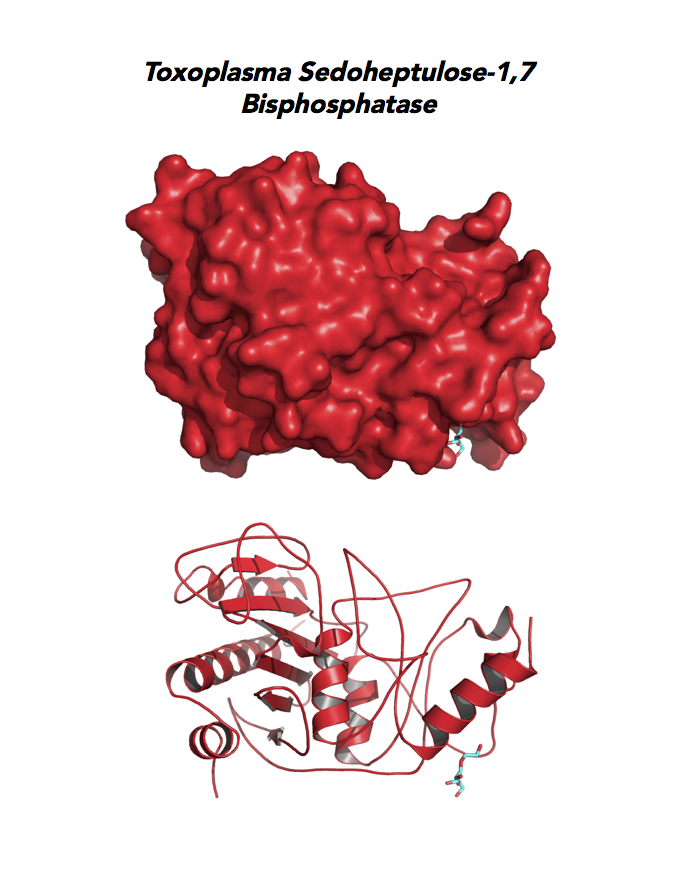
Target 7. Sedoheptulose-1,7 bisphosphatase (TgSBP).
SBP/SBPase regenerates 5-carbon sugars during the Calvin cycle in the chloroplast. The biphosphatases SBP and FBP/FBPase (fructose 1,6 bisphosphatase) have similar FBPase and AMP-binding domains, varying in their specificity for the 6- or 7-carbon substrates. Fructose bisphosphatase is a pivotal, anabolic enzyme in gluconeogenesis and the Calvin cycle.
FBP is a target for drug development to treat several cancers and type 2 diabetes. Second-generation FBP inhibitors targeting AMP binding have good results in clinical trials with non-human mammals and humans.
Toxoplasma expresses two FBPs and one SBP with substrate specificity in every stage of the parasite life cycle (unpublished data, David Roos' Laboratory). Essentiality and functions of these enzymes are not known. Structures of SBP and FBP from moss show similar redox-active regulatory disulfides mechanisms. They differ in surface areas of their active sites to accommodate specific substrate. As also found in Toxoplasma, SBP forms dimer and FBP assembles as a tetramer. Its essentiality, metabolic and moonlighting functions are not studied. Second-generation FBP inhibitors may be valuable drug leads in targeting TgSBP and TgFBPs.
PDB: 4IR8, 1.85Ao. UniProt: B9PW60. ToxoDB: TGME49_235700. EC#: 3.1.3.37. Molecular Function: Hydrolase Activity, Phosphatase Activity, Sedoheptulose Bisphosphatase Activity. Biological Process: Carbohydrate Metabolic Process, Dephosphorylation.
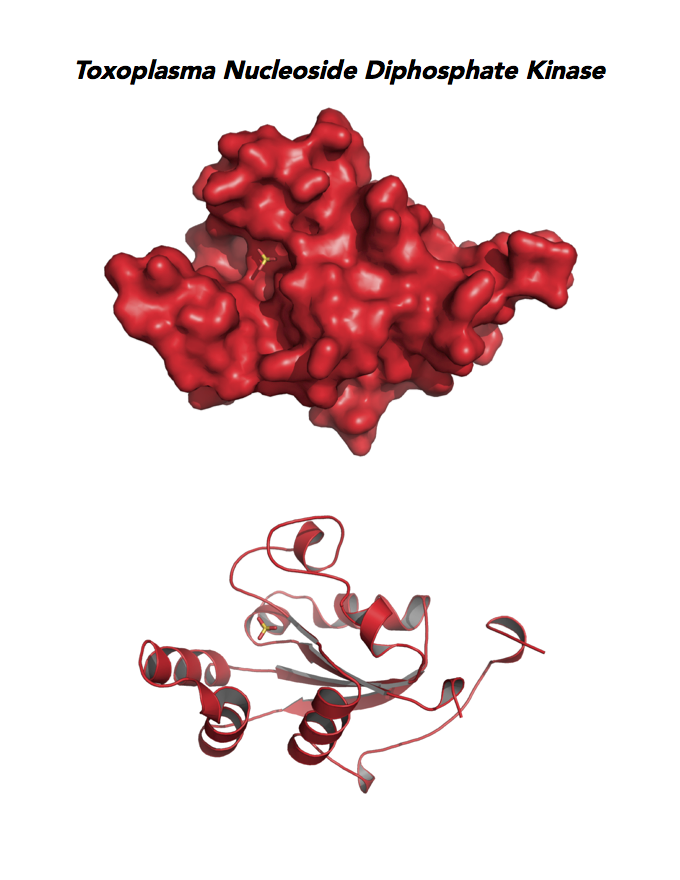
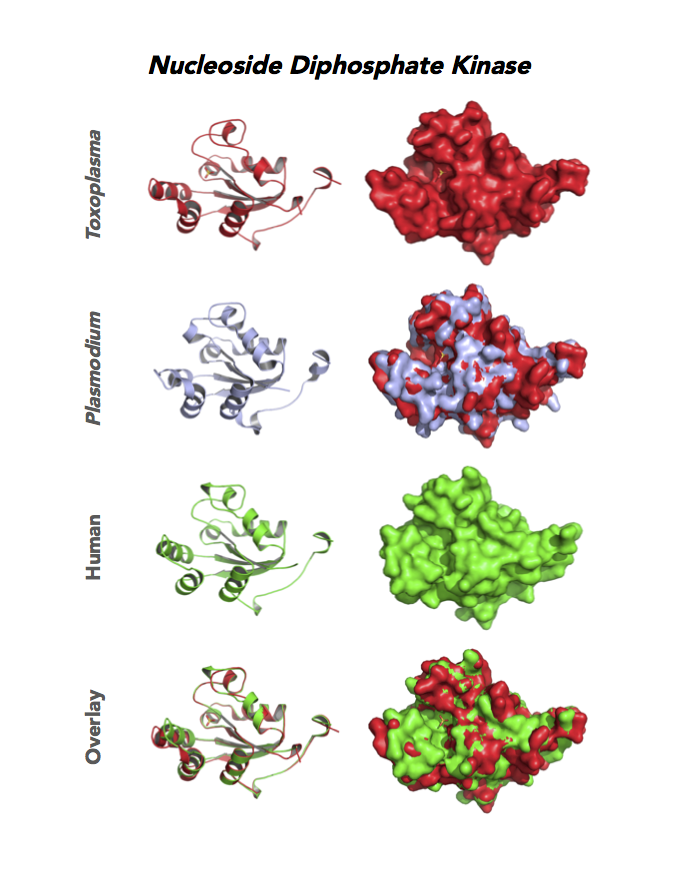
Target 8. Nucleoside diphosphate kinase (TgNDPK).
NDPK/NM23/NME is a multifunctional protein. NDPK generates nucleoside triphosphates important for nucleic acid synthesis, Krebs cycle, protein synthesis, G-protein signaling, and tubulin polymerization. NDPK also shows activities as histidine-dependent protein kinase and nuclease. This enzyme functions in many important processes: cell migration, growth and differentiation, signal transduction, transcriptional regulation, cytoskeleton regulation, apoptosis, embryonic development, neural activity, and skin homeostasis.
NDPK associates with cardiovascular disease and acts as a suppressor of most tumor metastasis, but pro-metastasis in neuroblastoma.
Both Toxo NDPKs express throughout the parasite life cycle, but this isoform upregulates sharply in the acute phase of mice infection and post-sporulation of oocysts (ToxoDB). This transferase may be important for purine salvage pathway from the host cell by this auxotrophic parasite. Its essentiality, metabolic and moonlighting functions are not known. TgNDPK structure allows comparison with human and Plasmodium NDPK/NM23 structures.
PDB: 4O0N, 2.4Ao, UniProt: S8FF85, ToxoDB: TGME49_295350, EC#: 2.7.4.6, Molecular Function: Nucleotide Binding, Nucleoside Diphosphate Kinase Activity, ATP Binding, Kinase Activity, Transferase Activity, Biological Process: Nucleoside Diphosphate Phosphorylation, GTP Biosynthetic Process, Utp Biosynthetic Process, Ctp Biosynthetic Process, Phosphorylation.
Figure 6. Eight panels show crystallographic structures of Toxoplasma enzymes, including Nucleoside diphosphate kinase (NDPK), Phosphoglycerate mutase (PGAM II), Orotidine 5'-monophosphate decarboxylase (OMPD), and Deoxyribose-phosphate aldolase-like (DPA).
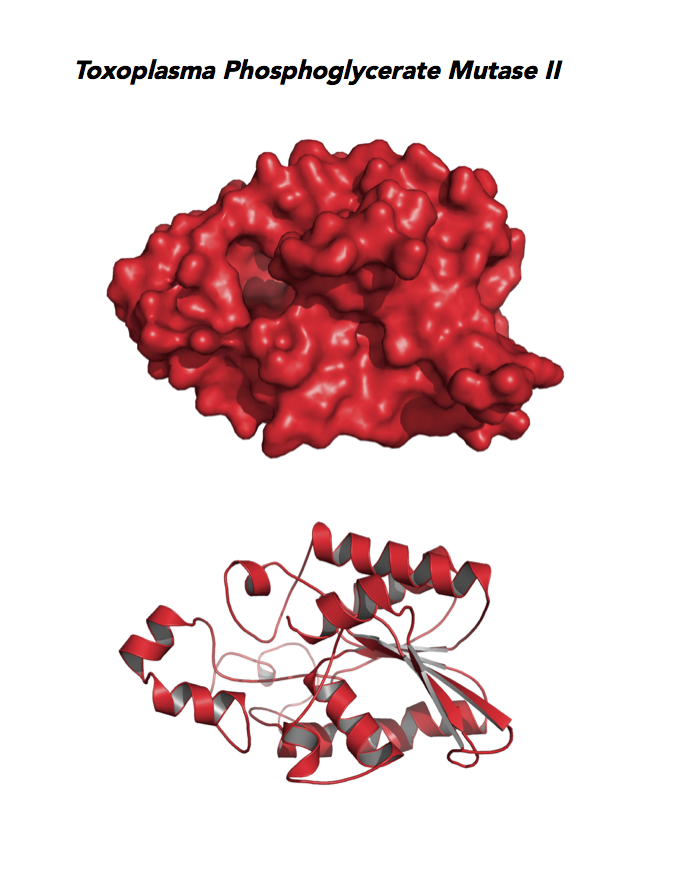
Target 9. Phosphoglycerate mutase II (TgPGAM II).
PGAM/PGM catalyzes step 8 of glycolysis. Cofactor-dependent type of PGM has three catalytic activities: mutase, phosphatase and synthase. In controlling intracellular levels of 3-PG and 2-PG, PGAM coordinates glycolysis, serine generation, and the oxidative arm of pentose phosphate pathway.
Genetic disorder of PGAM is mild to moderate and not thought as life threatening. The enzyme is upregulated and contributes to the Warburg effect in various human cancers, including breast, lung, prostate, and glioblastoma. Drug design is ongoing to find PGAM1 inhibitors to impede tumor growth. Multifunction is not examined.
Toxoplasma PGAM II is expressed across life cycle stages, but the isozyme is upregulated in the chronic phase of mice infection and the post-sporulation of oocysts in cats (ToxoDB). It is unclear whether TgPGAM II is targeted to the residual plastid. TgPGAM II shows potential as a vaccine candidate. TgPGAM II structure is most similar to the cofactor-dependent type of E. coli and human Type B PGAMs. Its essentiality, metabolic and moonlighting functions are not known. A structure of TgPGAM II allows comparison with human and Plasmodium analogues.
PDB: 4ODI, 2.6 Ao, UniProt: S8GJT7, ToxoDB: TGME49_297060, EC#: 5.4.2.4, Molecular Function: Catalytic Activity, Phosphoglycerate Mutase Activity, Intramolecular Transferase Activity Phosphotransferases, Biological Process: Glycolytic Process, Metabolic Process.
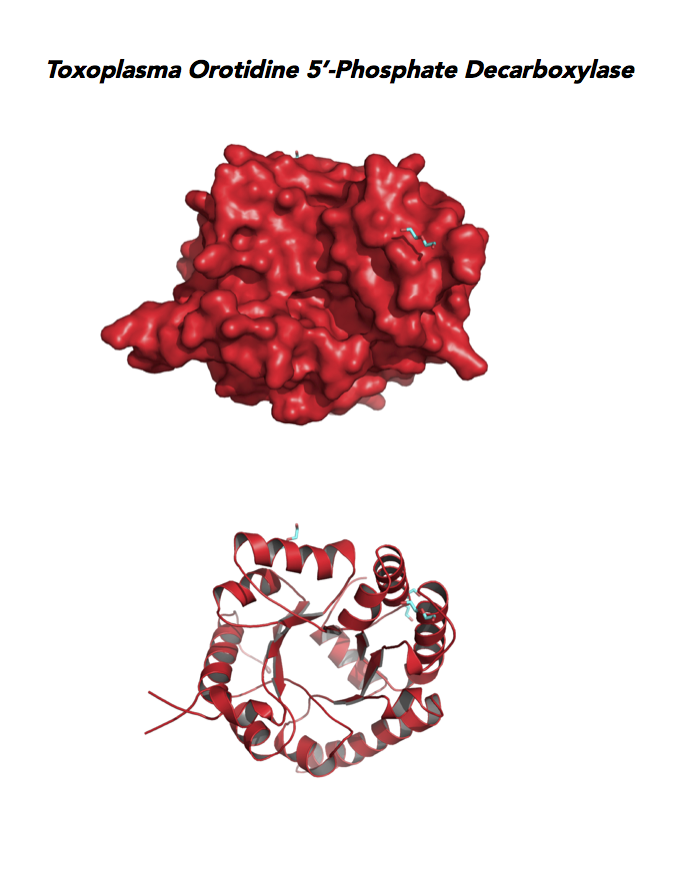
Target 10. Orotidine 5'-monophosphate decarboxylase (TgOMPD).
OMPD/ODCase catalyzes step 6 of de novo pyrimidine pathway. Pyrimidine nucleotides are essential precursors of RNA and DNA, cell membranes, protein glycosylation and glycogen synthesis. In human, step 5 and 6 are catalyzed by one bifunctional enzyme UMP synthase (UMPS), composed of orotate phosphoribosyltransferase (OPRT) and OMPD domains.
In Plasmodium, the native enzymes organize in an alpha2-beta2 heterotetramer structure having two subunits each of OPRT and OPD. De novo pyrimidine pathway links tightly to de novo folate biosynthesis in malarial parasites is missing in humans. Over 200 crystal structures of OMPD are reported, accompanied by a wide range of inhibitors.
Pyrimidine biosynthesis associates with the etiology, effect or treatment of many diseases (cancer, diabetes, AIDS and viral diseases, malaria and parasitic diseases, rheumatoid arthritis and autoimmune diseases). Moonlighting functions of enzymes in the de novo pyrimidine pathway are not studied.
Toxoplasma OMPD expresses in all life cycle stages, but the enzyme upregulates in the chronic phase of mice infection (ToxoDB). It shows high expression in oocysts, but downregulated in bradyzoite development and post-sporulation of oocysts. Its essentiality, metabolic and moonlighting functions remain unknown. TgOMPD structure allows comparison with human and Plasmodium analogues.
PDB: 4MJZ, 2.75Ao, UniProt: B6KBH9, ToxoDB: TGME49_259690, EC#: 4.1.1.23, Molecular Function: Catalytic Activity, Orotidine 5' Phosphate Decarboxylase Activity, Transferase Activity, Transferase Activity Transferring Glycosyl Groups, Lyase Activity, Biological Process: 'de Novo' Pyrimidine Nucleobase Biosynthetic Process, Pyrimidine Nucleotide, Biosynthetic Process, Metabolic Process.
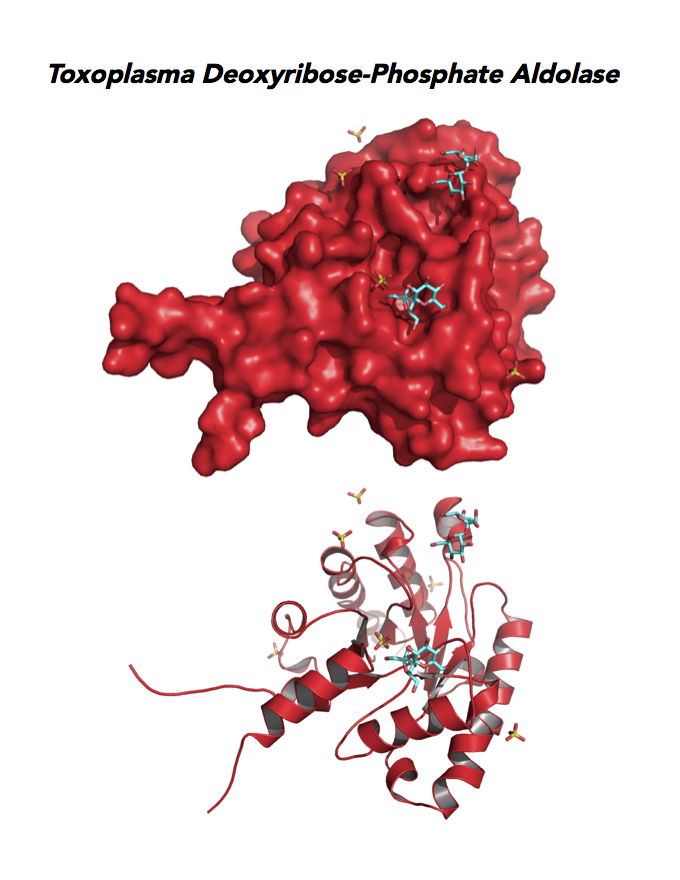
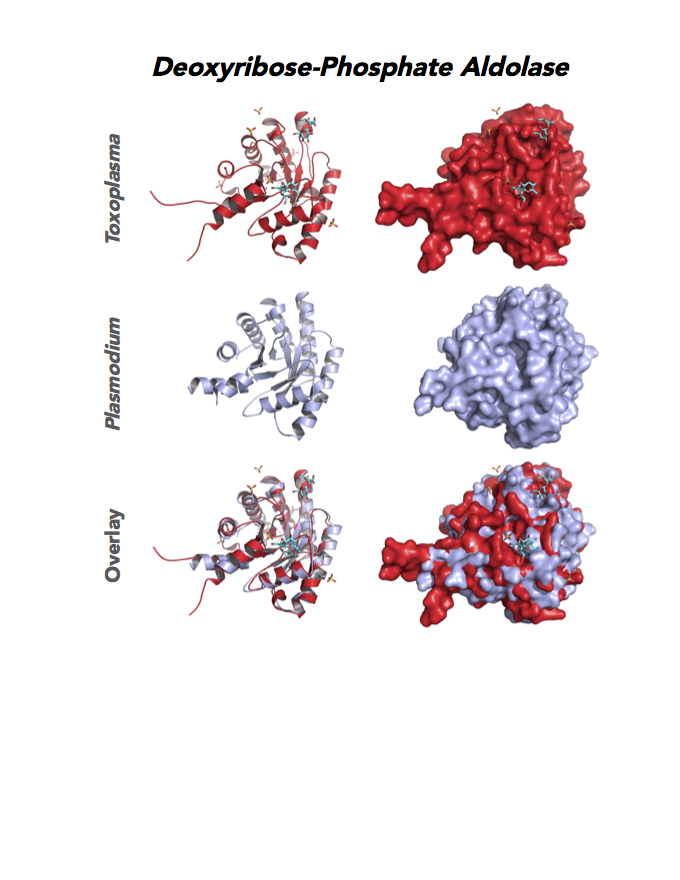
Target 11. Deoxyribose-phosphate aldolase-like (TgDPA).
Toxoplasma DPA expresses in the chronic infection of mice. The pseudoenzyme lacks the critical catalytic residues and is inactive. The protein scaffold likely serves an important role in brain cyst development. TgDPA dimer forms a cap-like structure that potentially binds to either polymerized actin, or TgADF (Actin Depolymerization Factor).
TgDPA may regulate actin filament dynamics and motility in the brain cyst. DNA vaccine encoding TgDPA (TGME49_270650) induce a strong humoral and cellular mediated (TH1, TH2 and TH17) response in mice. A combined approach of drug targeting both active enzyme and pseudoenzyme should be considered.
PMID: 25284756. Tonkin et al., 2015. J Mol Biol. 427:840-52.
PDB: 4EIV, 1.37Å, 3QYQ, 1.8Ao, UniProt: B9PVB4, ToxoDB: TGME49_118750, EC#: 4.1.2.4, Molecular Function: Deoxyribose Phosphate Aldolase Activity, Lyase Activity, Biological Process: Deoxyribonucleotide Catabolic Process.
Target 12. Ornithine aminotransferase (TgOAT).
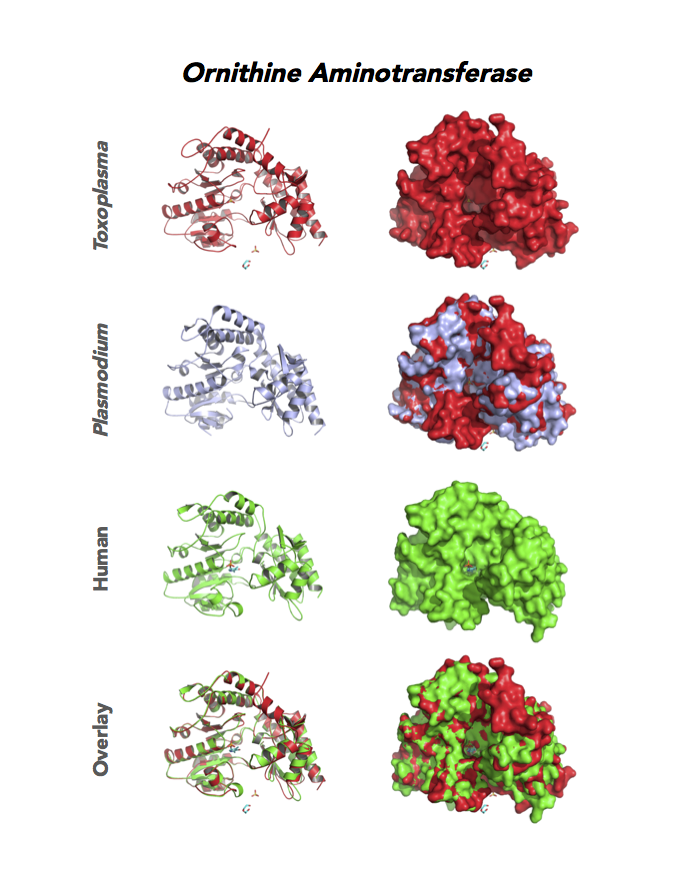
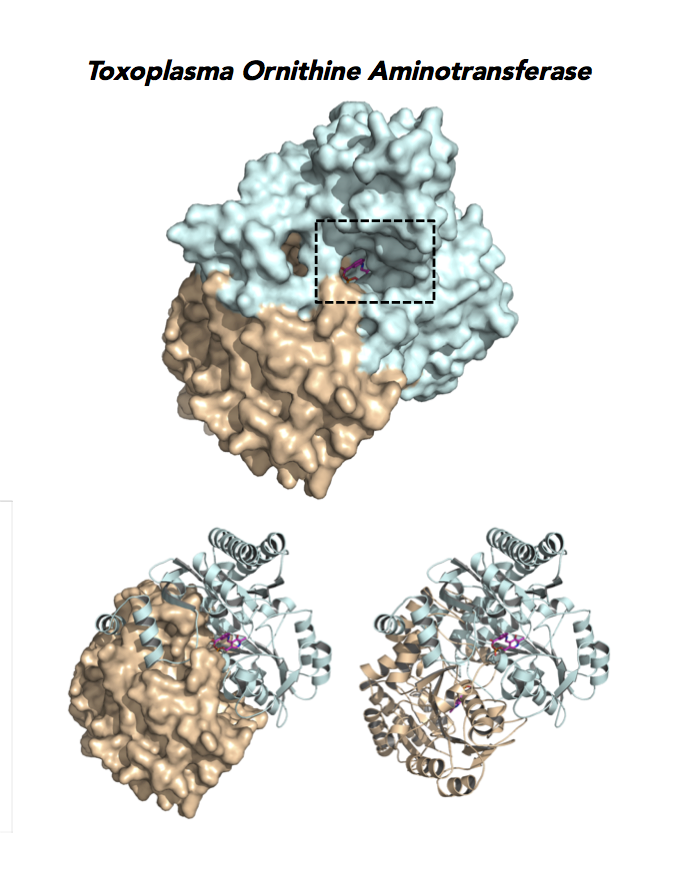
In mammals, OAT enriches in the liver, intestine, kidney and brain. The enzyme uses a cofactor PLP in a 2-step transamination to form glutamate from ornithine. The notable exception is the intestine, where citrulline and arginine are end products. OAT controls the production of signaling molecules and mediators, such as Glu, Cit, GABA, and aliphatic polyamines. It involves also in proline synthesis. Ornithine plays an important role of several metabolic diseases like hyperorithinemia, hyperammonemia, gyrate atrophy and cancer in humans
Toxoplasma is auxotrophic for polyamines. TgOAT expresses specifically in the oocyst/sporozoite stage of cat’s intestine. TgOAT uses the described transamination mechanism. The parasite enzyme however shows a dual N-acetyl-ornithine (AcOrn) and γ-aminobutyric acid (GABA) transaminase activity, due to Val79 in place of Tyr, as observed in mammalian homologues. TgOAT may function in both arginine and GABA metabolism. Structural and substrate differences in TgOAT make it an attractive target for chemotherapeutic intervention.
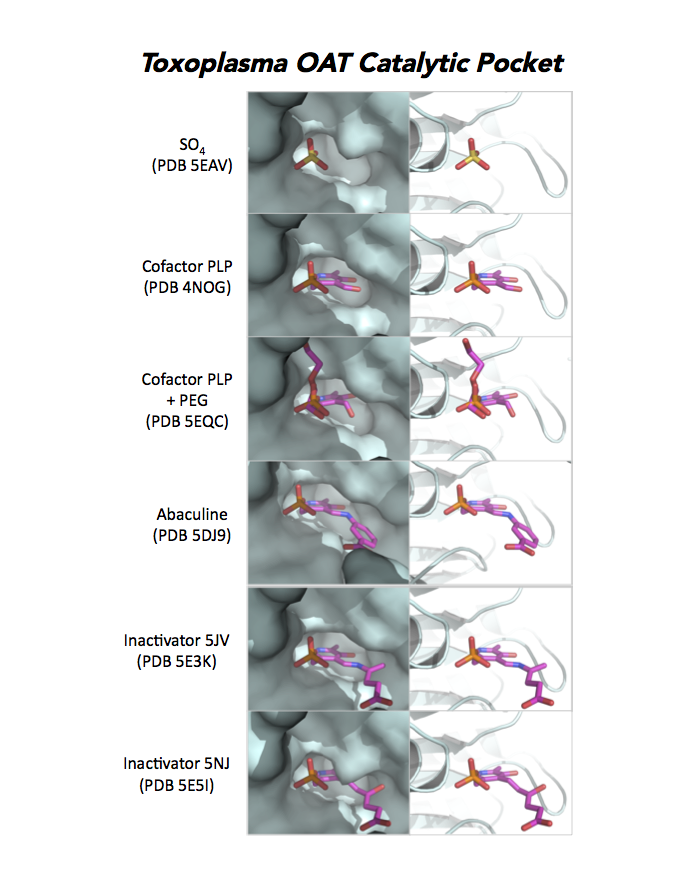
Dr. Richard Silverman's research group is working with CSGID to study the atomic resolution of ligands and prodrug binding to Toxoplasma OAT.
PDB: Unliganded, Apoenzyme (5EAV, 1.6 Å, 5UPR, 2.0 Å), Ligand: pyrodoxal-5'-phosphate, Holo-enzyme (4NOG, 1.2 Å, 4ZLV, 1.8 Å ; 4ZWM, 2.31 Å), Ligand: pyrodoxal-5'-phosphate, B- and D-Glucose (5EQC, 2.2 Å), Ligand: inactivator 5JV (5E3K, 1.7 Å), Ligand: inactivator 5JV, 5NJ (5E5I, 1.7 Å), Ligand: gabaculine (5DJ9, 1.55 Å).UniProt: S8EY38, ToxoDB: TGME49_269110, EC#: 2.6.1.13, 2.6.1.11, Molecular Function: Ornithine Oxo Acid Transaminase Activity, Transaminase Activity, Transferase Activity.
Figure 7. Three panels illustrate the current efforts to design drugs by studying the catalytic pocket of ornithine aminotransferase.